Foundations II: Technical Introduction to Pathogen Genomic Epidemiology: Mutations, Transmission, and Phylogenetics
- Continuing Education
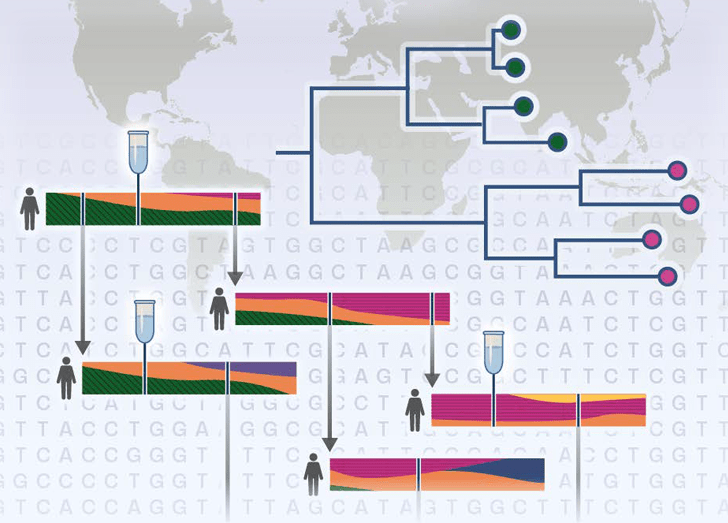
Explore how genomic patterns help us understand infectious disease transmission.
- Online; Self-Paced
This course is offered on demand and participants progress through the materials at their own pace.
Free
Continuing Education
2 Hours
On This Page
Overview
Foundations II: Technical Introduction to Pathogen Genomic Epidemiology: Mutations, Transmission, and Phylogenetics introduces learners to the molecular foundations underlying pathogen genomic sequencing.
The course explores how pathogens accumulate mutations as they replicate, creating distinct genomic patterns that can be leveraged to understand infectious disease transmission. You will examine the critical role of mutations in shaping genomic similarity, and how these patterns are visualized using phylogenetic trees to reconstruct relationships between infections. Building on this foundation, the course presents methods for integrating genomic and epidemiological data—including demographic details and contact tracing—to enhance outbreak investigations and support more informed public health decisions.
Key concepts are presented in brief, accessible video segments, with assessments throughout that allow you to check your understanding and apply your learning. With a focus on practical application and scientific principles, this course equips learners with the knowledge needed to inform effective disease surveillance and response in real-world public health contexts.
Learning Objectives
- Describe the underlying biological processes of pathogen replication, mutation, and transmission events.
- Explain how pathogen genomic sequences can provide insight into whether infections are likely to be closely linked within the chain of transmission.
- Interpret a phylogenetic tree to determine the relatedness between infections and make an epidemiological inference based on these data.
Who Should Enroll?
Foundations II: Technical Introduction to Pathogen Genomic Epidemiology: Mutations, Transmission, and Phylogenetics is open to any registrant and recommended for anyone involved in infectious disease surveillance or outbreak response from the domains of laboratory science, bioinformatics, or epidemiology. Some previous knowledge of genomics or molecular biology is beneficial but not required.
Completion of Foundations I is recommended but not required before taking Foundations II.
Course topics
Course Introduction
- Measles on the Move: Unraveling an Outbreak with Pathogen Genomics
The Biology Behind Pathogen Genomic Epidemiology
- Lesson Introduction
- Pathogen Transmission, Replication, and Mutations
- Reality vs. Data: What Sequencing Data Captures, Part 1
- Reality vs. Data: What Sequencing Data Captures, Part 2
Phylogenetics in Pathogen Genomic Epidemiology
- Lesson Introduction
- Introduction to Phylogenetic Trees
- Interpreting Phylogenetic Trees
- Navigating Different Types of Trees
- Phylogenetic Trees and Outbreak Response
Acknowledgments
These courses were developed under the U.S. Pathogen Genomics Centers of Excellence (PGCoE), and supported by the Office of Advanced Molecular Detection, U.S. Centers for Disease Control and Prevention (CDC) through Cooperative Agreement Number CK22-2204. Contents are solely the responsibility of the authors and do not necessarily represent the official views of the Centers for Disease Control and Prevention.
Course Directors

Pardis Sabeti
MD, DPhil
- Professor, Harvard T.H. Chan School of Public Health, Broad Institute of MIT and Harvard, Howard Hughes Medical Institute

Alli Black
PhD
- Senior Epidemiologist, Molecular Epidemiology Program, Office of Communicable Disease Epidemiology, Division of Disease Control and Health Statistics at Washington State Department of Health
"Capturing the nuances of this field isn’t easy, but this material offers an exceptional and friendly framework…useful for even the most knowledgeable genomic epidemiologists.”
Related Programs
Foundations I: Conceptual Foundations of Pathogen Genomics
Discover how pathogen genomics informs public health decisions and practice.
- Online; Self-Paced
Dates: Available Anytime
For: Anyone involved in infectious disease surveillance or outbreak response, as well as those in adjacent fields who would benefit from understanding the growing role of pathogen genomics in applied public health.
